Optimizing Protocols in K&A or Shasta Software
Optimizing your protocol can improve coupling efficiency, increase yields, reduce failed columns, and make your workflow more consistent
The single most important tip is:
Always watch 1 or 2 base cycle and adjust based on what you see.
No amount of protocol tweaking will help if you’re not visually confirming how your reagents behave during the run. Watching a cycle will tell you:
Whether deblock is fully removing trityl
If washes are clearing the previous reagent
If coupling looks strong and stable
If priming is working effectively (especially for Shasta)
Dedicate columns if needed to observe and optimize — it will save you time, money, and frustration later.
1. Coupling Times
Coupling time is a major factor in synthesis success:
-
DNA: Minimum 60 seconds
-
RNA: Minimum 6 minutes
-
Dyes / Complicated Monomers: 6 to 15 minutes
General rule: The greater the molecular size or steric hindrance, the more coupling time and possibly coupling repetitions you’ll need.
For longer oligos:
-
Add 1 extra coupling for every 40–60 bases. This can be done through the meta protocol
For large-scale syntheses (≥25 μmol):
-
Increase amidite concentration to maintain high coupling efficiency — especially for bulky or low-reactivity monomers.
2. Deblock
Deblock steps remove the trityl group from the 5’ end:
-
DCA: 15–60 seconds
-
TCA: 10–40 seconds
Tips:
-
Watch the cycle to ensure all trityl is removed.
-
Adjust deblock repetitions based on what you see.
-
For long oligos, add 1 extra deblock for every 40–60 bases.
3. Wash Steps
-
Washes can be short if they are enough to remove all traces of the previous reagent.
-
Watching the cycle will confirm this — don’t rely on time alone.
DEA Wash:
-
Use only in the final steps of synthesis.
-
Main purpose: Prevents N3 cyanoethylation.
-
Especially useful for RNA or sensitive modifications.
4. Sulfurization & Oxidation Order
Correct step order preserves backbone integrity:
-
Sulfurization: Always before capping — prevents acetic anhydride from interfering.
-
Oxidation: Best after capping. Add an extra capping step after oxidation to help remove moisture from oxidizer.
If using mixed sulfur and oxygen backbones:
-
PS → Cap → Ox → Cap
-
Oxidizer-only: Cap → Ox → Cap
-
Sulfur-only: Sulfur → Cap → Cap
5. Supports
Steric hindrance increases as the oligo grows:
-
Long oligos (60+ bases) / bulky groups : Use 2000 Å CPG for higher efficiency.
-
Polystyrene supports swell and are not recommended for K&A.
-
Standard support increases yield and does not require heat for cleavage.
💡 For standard supports:
A small percentage of Standard Support can become unfunctionalized. Add a capping step during initialization to block unlinked sites and prevent N-1 products — especially important with bulky 3’ modifiers.
6. Moisture Control
Even trace moisture reduces coupling efficiency.
-
Use fresh, anhydrous ACN and amidites.
-
Add trap packs (molecular sieves) to ACN, activator, and amidites at least 1 hour before synthesis.
-
Use in-line drying filters for argon/nitrogen.
-
Monitor room humidity near the instrument.
7. K&A vs Shasta Instrument Considerations
K&A – Trityl Monitor
-
Use trityl monitor to track coupling efficiency.
-
High initial V2 values can make readings unreliable — often a timing issue, not a chemistry failure.
-
Always confirm trityl data by watching the cycle.
- Check out our Understanding Trityl Monitor article for more information
Shasta – Crystallization & Priming
Shasta’s open column design makes crystallization common.
-
Prime infrequently used tips every every cycle — at least 1 prime, preferably 2.
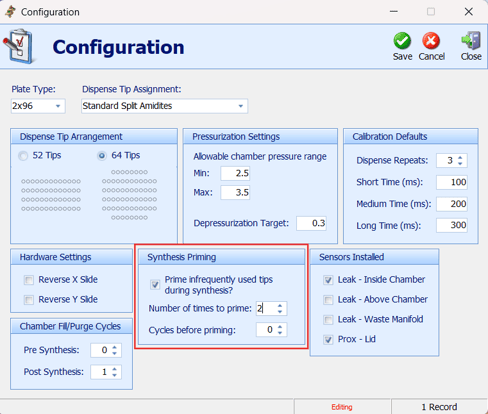
-
Ensure chamber is fully sealed to reduce solvent evaporation and crystallization.
-
O‑methyl‑U and LNA‑C: prone to crashing out. Use 10–20% DMF or THF as cosolvent.
-
Keep dummy “poly” sequences for routine priming (e.g., Poly O‑methyl‑U) — ensures amidites are primed every cycle, reducing random column failures.
8. Saving Protocol Versions
-
Each change should be saved as a new protocol (e.g., RNA_Long_v2).
-
Document what was changed and why.
-
Compare results to your baseline runs.
When to Contact Support
If, after optimization, you still see:
-
Severe yield loss
-
Inconsistent coupling
-
Random unexplained failures
Contact our service team with your run files and protocol settings.
